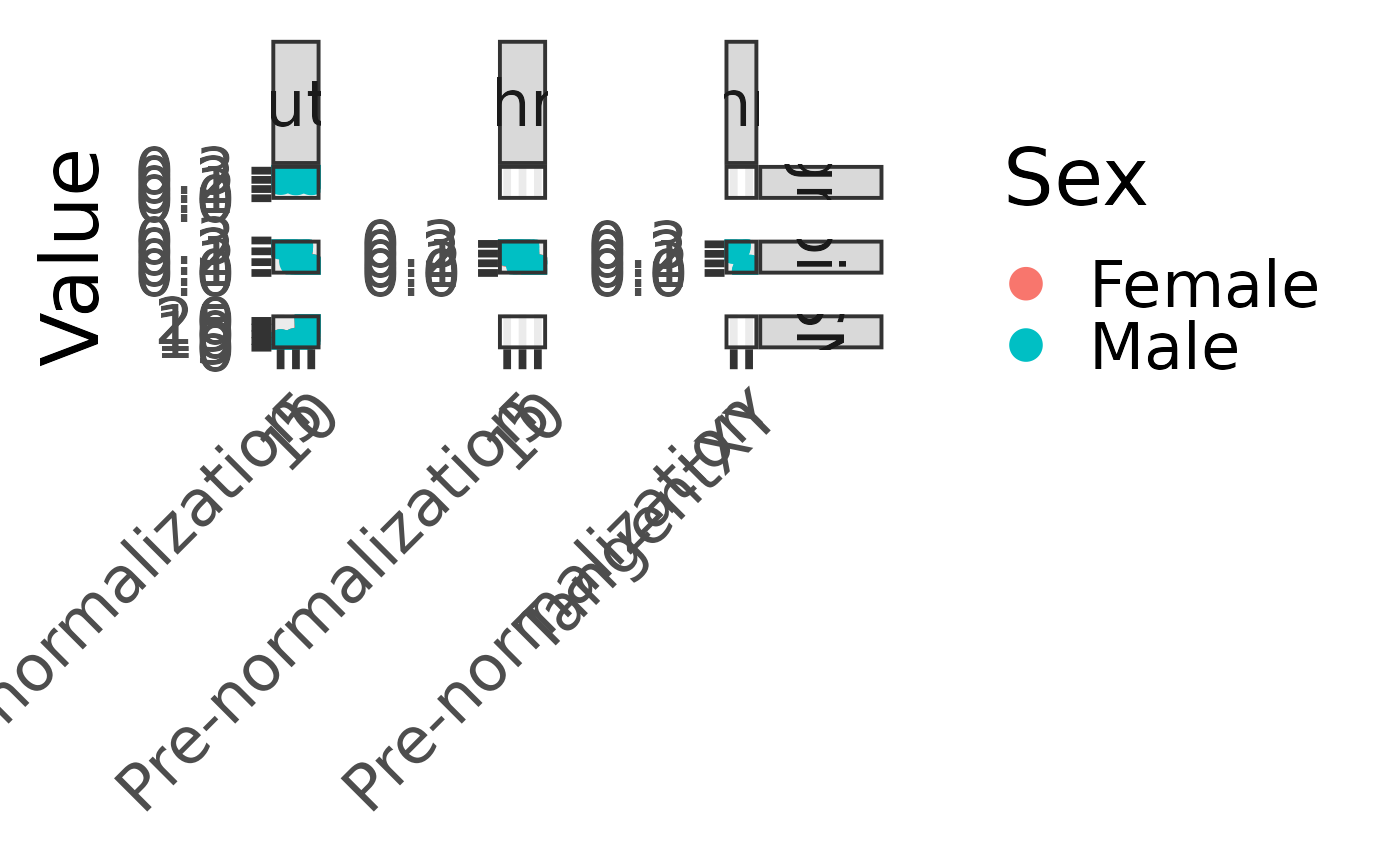
Plot signal and noise before and after TangentXY normalization
plot_signal_noise.RdThis function calculates and plots the signal and noise profiles of tumor samples before and after TangentXY normalization with varying numbers of latent factors. The signal is defined as the standard deviation of the median signal of each chromosome arm (grouped into autosomes, X, and Y chromosomes), and the noise is defined as the median of the absolute values of the vector of consecutive differences between each signal.
Arguments
- tnorm
A normalized tumor signal matrix output by run_tangent, or a list of such matrices.
- sif_df
Tibble or filepath to a text file containing sample metadata
- pif_df
Tibble or filepath to a text file containing probe information. Should have a 'chr' column with chromosome names (1-22, X, Y) and an 'arm' column with arm names (p or q). Should also have a 'locus' column with locus names in the format
"{chr}:{start}-{end}"where{chr}is 1-22, X, or Y and where{start}and{end}are genomic coordinates.- tsig_df
Tibble or filepath to a text file containing the tumor signal matrix
- n_latent
A numeric vector of the numbers of latent factors used in TangentXY normalization. Should correspond to the elements in
tnorm.- output_dir
Directory to save the plot. If
NULL, the plot will be printed to the screen.- n_cores
Number of cores to use for parallel processing.
Value
(Invisibly) A tibble containing the signal and noise profiles for each sample before and after normalization.
Examples
n_latent <- c(5, 10)
tangent_res <- lapply(n_latent, function(nlf) {
run_tangent(example_sif, example_nsig_df, example_tsig_df, nlf, make_plots = FALSE)
})
#>
#> Applying linear transformation ...
#>
#> Running SVD ...
#>
#> Running Tangent on autosomes and chrX ...
#> Done.
#>
#> Running Tangent on male chrY ...
#> Done.
#>
#> Applying linear transformation ...
#>
#> Running SVD ...
#>
#> Running Tangent on autosomes and chrX ...
#> Done.
#>
#> Running Tangent on male chrY ...
#> Done.
res <- plot_signal_noise(tangent_res, example_sif, example_pif,
example_tsig_df, n_latent = n_latent)
#> Calculating signal and noise in pre-normalization data...
#> Calculating signal and noise in normalized data with 5 latent factors...
#> Calculating signal and noise in normalized data with 10 latent factors...
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to max; returning -Inf
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to max; returning -Inf
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to max; returning -Inf
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to max; returning -Inf
#> Warning: no non-missing arguments to min; returning Inf